Download PDF format
A BIOINFORMATICS CHARACTERIZATION OF THE RNA-DEPENDENT RNA POLYMERASE FROM THE PARASITIC FUNGI, Metarhizium anisopliae
C. N. WEIR1, 2 M. Mol. Biol. and Ph. D candidate
E. WONG Ph. D. 3
G. F. KING Ph. D.4
1 Walter and Eliza Hall Institute of Medical Research, 1G Royal Parade, Parkville 3052, Victoria, Australia, e-mail: weir.c@wehi.edu.au
2 University of Melbourne, Department of Medical Biology, Parkville 3010, Victoria, Australia
3 European Bioinformatics Institute (EMBL-EBI), Wellcome Trust Genome Campus, Hinxton, Cambridgeshire CB10 1SD, United Kingdom
4 Institute for Molecular Bioscience, University of Queensland, St. Lucia 4072, Brisbane, Queensland, Australia
Metarhizium anisopliae is an entomopathogenic organism that is parasitic towards insects, including those that serve as vectors for human disease, including malaria parasite infected mosquitoes. Directed bio-engineering of parasitic has been proposed as a potentially novel vector control strategy. However, many parasitic fungi posses a RNA-dependent RNA polymerase () gene that serves as a defence against transgenes. Here we characterize the gene of , including the context of the gene in relation to surrounding genes and theoretical modeling based on the RdRp enzyme of . AnophelesM. anisopliaeRdRpRdRpM. anisopliaeNeurospora crassa
Keywords: entomopathogenic fungi, Metarhizium anisopliae, parasites, transgenes, vector control.
In fungi, including parasitic Cordyceps species and some of the Metarhizium species, RNA-dependent RNA polymerases (RdRp) are involved in the phenomenon of gene-silencing, or quelling, where they act at the post-transcriptional level, affecting both endogenous genes and transgenes1. Slight differences in molecular pathways exist depending on what type of gene is involved, however here we will focus on exogenous transgenes (for a review of both mechanisms, please refer to Dang Y et al.2). During quelling, a diffusible trans-acting aberrant RNA (abRNA) molecule of the expressed gene is produced and converted to double-stranded RNA (dsRNA) via RdRp’s activity allowing for the synthesis of complementary RNA molecules1-3. The dsRNA is recognized by the Dicer enzyme (or Dicer-like proteins), degrading the dsRNA into small-interfering RNA (siRNA) molecules that get incorporated into the RNA-induced silencing complex (RISC)1,2. RISC uses the siRNA’s to guide nuclease activity against normal messenger RNA (mRNA) leading to degradation and thus gene silencing1,4.
Transgene incorporation into fungi does not appear to be enough to trigger RdRp’s activity; tandem repeats of transgenes, however, results in quelling2. This is important, because of research to genetically engineer an entomopathogenic parasitic fungi, Metarhizium anisopliae, with various transgenes, including spider-venom peptides, for use as a disease vector control agent, such as malaria-spreading Anopheles mosquitoes (see references for examples of prior experiments and toxin information)5-8. Previously it was shown that Metarhizium strains bio-engineered with the scorpion toxin scorpine and an tandem 8-fold repeat of salivary gland and midgut peptide 1 (SM1; blocks Plasmodium sporozoites entrance into mosquito salivary glands) both reduced sporozoite density by over 95 % and killed all Plasmodium parasite-infected mosquitoes by days 14–17 and 20 respectively6. Whilst gene silencing, if present, did not appear to be problematic, there is no guarantee that future transgene tandem arrays will be expressed successfully6. Additionally, activity may have been improved if quelling was prevented. Due to this, a bioinformatic analysis of M. anisopliae’s genome (solved in 2011)9 has been carried out to search for RdRp homologues. This analysis includes gene structure and context, domain organization, homology and phylogenetics for RdRp, plus the use of gene prediction software and analysis for previously unannotated genes near RdRp’s location. Finally, this paper examines the likely structure of the catalytic site of M. anisopliae RdRp based on homology with an existing RdRp10.
Materials and methods
Gene structure and context
The annotated RdRp gene from M. anisopliae strain ARSEF23 was found in the EMBL-EBI webpage (http://www.ebi.ac.uk/ena/) (Accession EFZ03903.1). The promoter region was identified using Transfac’s Patch 1.0 (www.gene-regulation.com/pub/programs.html) using the 200 nucleotides upstream from the RdRp gene as an input. Distances between genes were confirmed as appropriate using information from the reference cited13.
The context of the RdRp gene context was determined using data obtained from the previous EMBL-EBI search (http://www.ebi.ac.uk/ena/data/-view/EFZ03903) and an additional search for the contig code GL698711.1. Genes found on c234 were found and their locations elucidated using NCBI’s nucleotide search (Accession GL698711.1). Reverse complement carried out using software found at http://www.rcsb.org/pdb/explore/explore.do?structureId=2J7N. This information allowed for distances and locations of surrounding genes to be elucidated. The additional gene context information was obtained by using the above data and Softberry’s Fgenesh gene search tool (http://linux1.softberry.com/berry.phtml?topic=fgenesh HYPERLINK "http://linux1.softberry.com/berry.phtml?topic=fgenesh&group=programs&subgroup=gfind"& HYPERLINK "http://linux1.softberry.com/berry.phtml?topic=fgenesh&group=programs&subgroup=gfind"group=programs HYPERLINK "http://linux1.softberry.com/berry.phtml?topic=fgenesh&group=programs&subgroup=gfind"& HYPERLINK "http://linux1.softberry.com/berry.phtml?topic=fgenesh&group=programs&subgroup=gfind"subgroup=gfind).
Protein Blast and Domain Structure
Protein blast searches were performed using the NCBI BLASTp function; BLAST searches were first performed only against fungi then against solved PDB entries (with the latter generating the information regarding Neurosporacrassa). Domain architecture information was generated with both NCBI (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi?;and and Pfam (Pfam.sanger.ac.uk/protein/E9EKW8). The sequence alignment between N. crassa and M. anisopliae RdRp was manually edited in order to improve the alignment. This was checked with the associated PDB file for PBD accession 2J7N.Pymol software used for modeling.
Homology
Fungi analyzed were selected based upon BLASTp results and those related in the literature9. For protein selection, the highest E-value results were used and hypothetical and predicted proteins were excluded unless, upon analyzing their information on NCBI, they showed evidence for being a true RdRp, such as similar sequences. Multiple sequence alignments were performed using Muscle (http://www.ebi.ac.uk/Tools/msa/muscle/). This information was used with MEGA 5.05 software including Muscle alignments, and phylogenetic tools. A phylogenetic neighbour joining tree was generated with the Bootstrap method (500 bootstraps used), Poisson modeling and pair wise deletion for the data treatment and amino acid substitutions allowed in the model. It should be noted that whilst M. acridium is highly related to M. anisopliae, it was excluded from the phylogenetic analysis of RdRp since the sequence that currently exists for it is incomplete (accession EFY87605).
Results and discussion
Gene Structure
The genome and transcriptome of M. anisopliae had been sequenced by Gao Q et al.9 and genes curated using EMBL-EBI software. The genome was shotgun-sequenced into 1271 contigs with each loci tagged as MAA; the RdRp gene (MAA_00977) is located on contig ADNJ01000234.1 (c234)9. After searching EMBL-EBI for «M. anisopliae ARSEF 23 RNA-dependant RNA polymerase» relevant gene information was obtained. Sequence length was reported as 3345 base pairs (minus the 222bp of 3 introns), comprised of 4 exons corresponding to the predicted 1114 amino acids. The transcriptional start site is coded for by amethionine, and the stop codon sequence is TAG. No promoters were described, so an attempt was made to find possible ones using Transfac (see Methods). A FASTA formatted sequence 200 bpupstream (towards the 5’ region) of RdRp’s start codon was input to the Transfac’s Patch 1.0 tool, which uses an algorithm for pattern-based prediction of transcription factor binding sites. A likely promoter site, GAGTCA, was found to be 31 bpupstream of the aforementioned start codon. With a score of 100 and no mismatches, this corresponded to a region where the binding factor characterized in Saccharomyces cerevisiae, GCN4, interacts.
Gene Context
Currently, the chromosome location of RdRp is unknown; only the contig location c234 exists. Gene prediction software (Fgenesh by Softberry) was used to predict adjacent genes along c234. EMBL-EBI software gave information pertaining to locus tags (identifiers which are applied systematically to all genes in a genome within the context of sequencing projects) for genes within each contig. Using this, the overall context of RdRp and its surrounding characterized genes could be viewed, along with contig assembly. The two closest downstream genes from RdRp’s stop codon, reported from the gene organization within the contigs, are those coding for the C6 transcription factor GliZ2 and a protein efflux pump, which are located 6477 and 12638 nucleotides away respectively. The nearest upstream gene, which is 9885 nucleotides away from the start codon of RdRp, codes for a hypothetical protein.
Given the distance from RdRp, both for upstream and downstream genes, a manual search for closer genes was carried out. As Figure 1 reveals, two such genes were identified either side of RdRp; a peroxidase protein 1943 nucleotides upstream of RdRp’s start codon, and the NFX1-type zinc finger-containing protein 1, which us 569 nucleotides downstream of RdRp’s stop codon. This figure, approximately to scale, shows the improved gene context of RdRp with respect to its surrounding genes after manually searching. Directionality of transcription is shown as arrows above the genes, with their size (including introns) and nucleotide location on the contig given below.
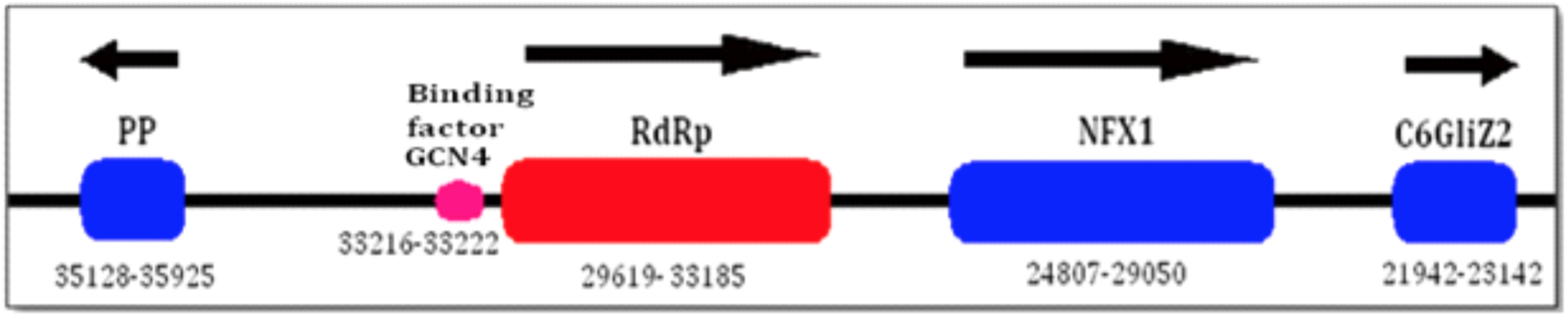
Figure 1. Gene context of RdRp:
PP – peroxidase protein; NFX1 – NFX1-type zinc finger-containing protein. All gene sizes shown are inclusive of introns and exons
Protein Blast and Domain Structure
A search of the conserved domain database at NCBI and the Pfam database using the amino acid sequence of RdRp, was performed (see Methods). The results are shown in Figure 2A and 2B. Both NCBI and Pfam revealed a single characterized domain for RdRp, corresponding to amino acid residues 314–925. The Pfam database indicates this family of proteins are important for eukaryotic post-transcriptional gene silencing and that there is a core catalytic domain responsible for the activity. Comparisons of the sequence similarity between this domain and the RdRp domain in the orthologous protein from the fungus Neurosporacrassa. N. crassa was used for two reasons; firstly, the RdRp of N. crassa has been well characterized including a complete crystal structure10. Secondly, as Gao Q et al. shows9, they are both related at the class level. There is 30 % identity between a 333-residue stretch within the given domain and the active catalytic sub-domain from the N. crassa protein (Figure 2C).
Despite low overall sequence homology, the catalytic aspartate residues are conserved between the two species10,11. The catalytic aspartate residues interact with a magnesium cation and are responsible for the selection of ribonucleoside triphosphates incorporated into the incoming abRNA, making it dsRNA10,12. These conserved aspartate residues are shown in Figure 2C at residues 1007, 1009 and 1011 in the sequence of N. сrassa RdRp.
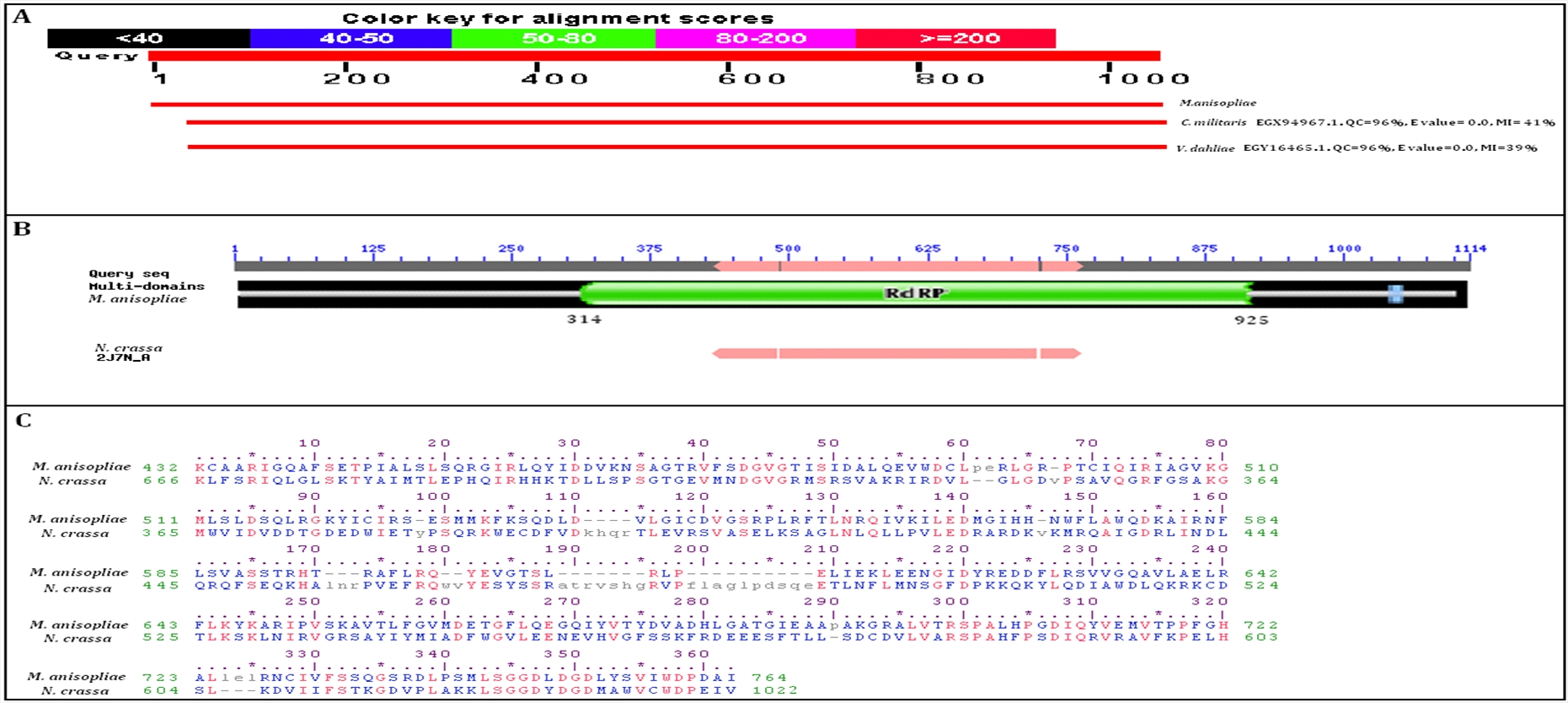
Figure 2. BLASTp and domain information:
A – The top two hits after blasting M. anisopliae RdRp against known fungal proteins. RdRp proteins from Cordyceps militaris and Verticillium dahlia showed highest sequence coverage and maximum sequence identity to M. anisopliae RdRp;
B – Domain architecture of M. anisopliae RdRp showing the 611-residue domain, compared to the catalytic sub-domain of N. crassa RdRp. The figure was made using both NCBI and Pfam data;
C – Sequence alignment of residues from (B) showing that M. anisopliae RdRp has 30 % sequence identity over 333 residues to N. crassa RdRp with an E-value of 1.34e-18. Residues in orange are homologous
Homology
The RdRp gene is found in viruses, plants, protozoa, fungi, and nematodes, although it is absent in insects or vertebrates. There are differences among these organisms with respect to domain organization which results in a catalytic domain possessing either right handed or double-barrelled structural organization10. Fungi, in contrast to viruses, possess the latter organization10, which will is discussed below in the Additional Information section. Figure 3 shows the phylogenetic relationship between various fungal species and their RdRp protein. The organization of fungi shown in Figure 3 corresponds to the published phylogenetic tree (see Gao Q et al.9), showing inferred evolutionary relationships between each fungus.
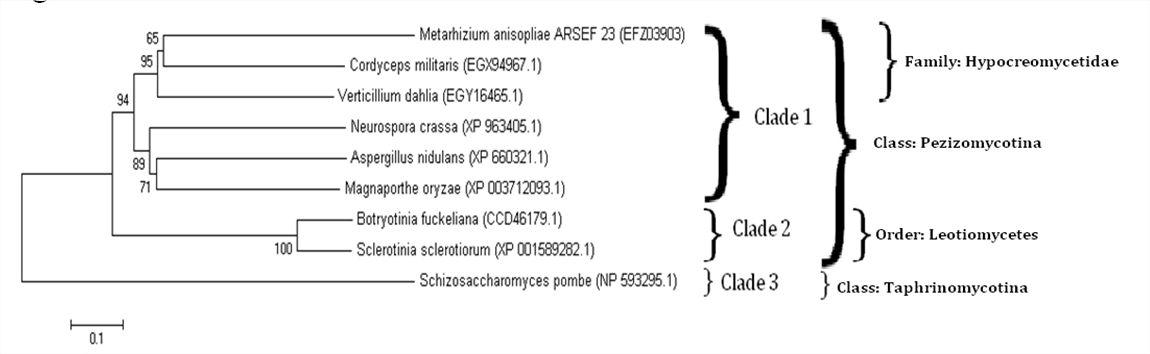
Figure 3. Phylogenetic relationships between fungal RdRps
This neighbour-joining tree, made using MEGA 5.05 (see Methods), demonstrates the evolutionary relationships between nine fungal RdRp proteins based on amino acid sequences. There are three main clades, which match overall the organization seen amongst genomes of the same fungi9. Clade 1 contains M. anisopliae, C. militaris, V. dahlia, N. crassa, A.nidulans and M. oryzae. Clade 2 contains B. fuckeliana and S. sclerotiorum. Clade 3 contains S. pombe. Taxonomic grouping is also demonstrated. The cut-off for significance regarding bootstrap values was anything below 65. The Poisson model was used in MEGA 5.05’s phylogeny tool and 500 bootstraps were selected.
Visualization of the Catalytic Region of RdRp
No 3D structure currently exists for M. аnisopliae RdRp, however there is one for N. crassa RdRp, as shown on the PDB under the identifying number 2J7N. It was decided, given the reasonable level of sequence identity between the two sequences shown in Figure 2C, that identification of the residues in the catalytic region of M. anisopliae RdRp should be carried out (Figure 4). This revealed the catalytic site with the predicted «catalytic» arginine residues shown interacting with a magnesium cation. These results show that all of the key residues reported in the literature with respect to the catalytic activity of N. crassa RdRpare conserved in the RdRp of M. anisopliae. The tyrosine residue at position 1010, which is sandwiched between the catalytic residues D1009 and D1011 in N. crassa RdRp (Figure 4), is substituted for a leucine in M. anisopliae RdRp. Given that both of these residues possess relatively large hydrophobic side-chains, it is unlikely that this substitution would have any negative effects on catalytic activity.
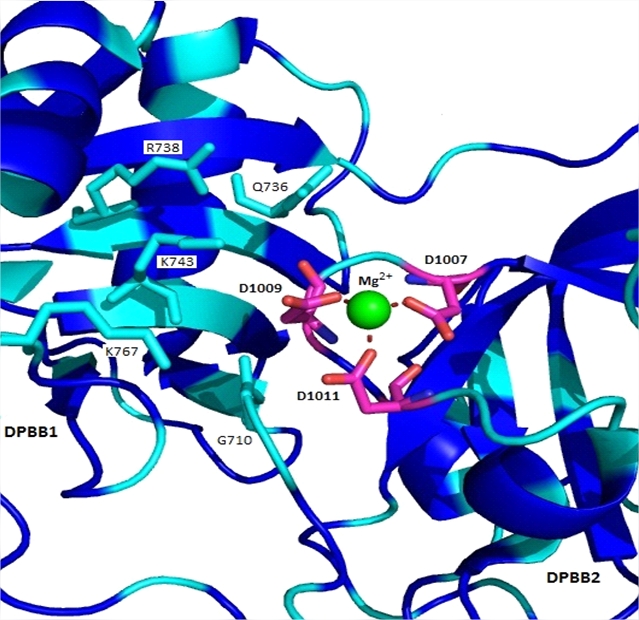
Figure 4. Catalytic site of RdRp
The crystallographic model solved to 2.3 Ǻ of N. crassa RdRp was used as the model of homology (PDB under accession number 2J7N). Key residues that are highly conserved among all known RdRps are labeled, and those that are identical in M. anisopliae RdRp are colored cyan (the blue are found in N. crassa RdRp).The three homologous catalytic aspartic acid residues are labeled and colored pink, and are shown interacting with the magnesium cation that lies between each DPBB. There is a tyrosine residue at amino acid position 1010 (not labeled, but is shown in blue) which in the corresponding sequence in M. anisopliae is a leucine. The distance of the interaction between each catalytic aspartic acid residue and the magnesium cation is between 2.1 and 2.2 Ǻ.
RdRp enzymes are found in a variety of both eukaryotic and prokaryotic organisms and serve as regulators of gene expression at the post-transcriptional level10-12. Given our future research directions involving transgene incorporation into M. anisopliae to improve its efficacy for controlling insect vectors of human disease, and the fact that transgenes have been known to initiate gene silencing, it is important to characterize the mechanisms behind this2,5,6. This report has elaborated on the gene silencing protein, RdRp, in M. anisopliae via the use of bioinformatic tools, with several key findings made.
During RdRp gene characterization, the finding that the promoter site GAGTCA is the sequence for binding factor GCN4 was of interest. GCN4 has been associated with the activation of genes involved in protein and purine synthesis, and expressed during times of stress14. One could consider the introduction and expression of transgenes to be a stressor to cells since indeed this has been reported in plants with similar silencing mechanisms to fungi, which evolved to possess viral defence mechanisms1,15. It therefore makes sense that this region would lie near RdRp.
Despite the fairly low overall sequence identity of 30 % between M. anisopliae and N. crassa RdRp, the conservation of key residues in the catalytic site was expected as predicted from the literature10,11. Modeling and matching identical residues was a good way of showing this and relating the form to function (Figure 4). The sequences lying outside of the domain given in Figure 3B for M. anisopliae RdRp may code for areas of the protein such as the slab, head or neck regions of the full protein.
The identification and analysis of the M. anisopliae RdRp gene reveals that there is likely to be a fully functioning molecular mechanism involved in gene silencing in M. anisopliae. This paves the way for development of knockout strains (such as the qde-1 knockout of N. crassa) that may have improved transgene expression3. The time taken (20 days) for the transgenic Metarhizium engineered by Fang et al. to kill Plasmodium-infected mosquitoes may have been reduced with the use of a RdRp knockout, if indeed transgene silencing was occurring6.
In the future, fluorescent in situ hybridization could be used to locate the chromosome on which RdRp is found16. Additionally, it would be of interest to work out the crystal structure of M. anisopliae RdRp since it is gaining popularity for use as an entomopathogenic agent.
Literature
1. Nakayashiki H. RNA silencing in fungi: Mechanisms and applications // FEBS Letters. – 2005. – V. 579. – P. 5950–5957.
2. Dang Y. et al. RNA Interference in fungi: Pathways, functions and applications // Eukary Cell. – 2011. – V. 10. – P. 1148–1155.
3. Cogoni C., Macino G. Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependant RNA polymerase // Nature. – 1999. – V. 399. – P. 166–169.
4. Cogoni C. Homology-dependant gene silencing mechanisms in fungi // Annu. Rev. Microbiol. – 2001. – V. 55. – P. 381–406.
5. Fang W., Azimzadeh P., St. Leger R.J. Strain improvement of fungal insecticides for controlling insect pests and vector-borne diseases // Curr. Opin. Microbiol. – 2012. – V. 15. – P. 1–7.
6. Fang W. et al. Development of transgenic fungi that kill human malaria parasites in mosquitoes // Science. – 2011. – V. 331. – P. 1074–1077.
7. Wang C., St. Leger R.J. A scorpion neurotoxin increases the potency of a fungal insecticide // Nature Biotech. – 2007. – V. 25. – P. 1455–1456.
8. Saez N.J. et al. Spider-venom peptides as therapeutics // Toxins. – 2010. – V. 2. – P. 2851–2871.
9. Gao Q. et al. Genome sequencing and comparative transcriptomics of the model entomopathogenic fungi Metarhizium anisopliae and M. acridium // PLoS Genetics. – 2011. – V. 7. – P. 100–126.
10. Salgado P.S. et al. The structure of an RNAi polymerase links RNA silencing and transcription // PLoS Biol. – 2006. – V. 4. – e434.
11. Gohara D.W. et al. Poliovirus RNA-dependent RNA polymerase (3Dpol): structural, biochemical, and biological analysis of conserved structural motifs A and B // J. Biol. Chem. – 2000. – V. 275. – P. 25523–25532.
12. O'Reilly E.K., Kao C.C. Analysis of RNA-dependent RNA polymerase structure and function as guided by known polymerase structures and computer predictions of secondary structure // Virology. – 1998. – V. 252. – P. 287–303.
13. Kupfer D.M. et al. Introns and splicing elements of five diverse fungi // Eukary Cell. – 2004. – V. 3. – P. 1088–10100.
14. Mascarenhas C. et al. Gcn4 is required for the response to peroxide stress in the yeast Saccharomyces cerevisiae // MBoC. – 2008. – V. 19. – P. 2995–3007.
15. Vance V., Vaucheret H. RNA silencing in plants – defense and counter defense // Science. – 2001. – V. 292. – P. 2277–2280.
16. Cremer T., Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells // Nature Rev Genetics. – 2001. – V. 2. – P. 292–302.
© 2016 The Author(s). Published by All-Russian Scientific Research Institute of Fundamental and Applied Parasitology of Animals and Plants named after K.I. Skryabin.
This is an open access article under the Agreement of 02.07.2014 (Russian Science Citation Index (RSCI) and the Agreement of 12.06.2014 (CABI.org / Human Sciences section).